‘This is a sustainable and straightforward approach to the problem of antibiotic resistance,’ said Fredrik Almqvist, professor of organic chemistry at Umeå University in Sweden. ‘New antibiotics take a huge amount of money to develop. We are developing compounds that boost the antibiotics we already have, which is much more cost effective.’
These new compounds don’t kill bacteria. Instead, they selectively disarm them, giving antibiotics the chance to swoop in and finish the job. ‘They sensitise the pathogen and restore the effectiveness of antibiotics,’ said Prof. Almqvist. ‘It’s amazingly cool.’
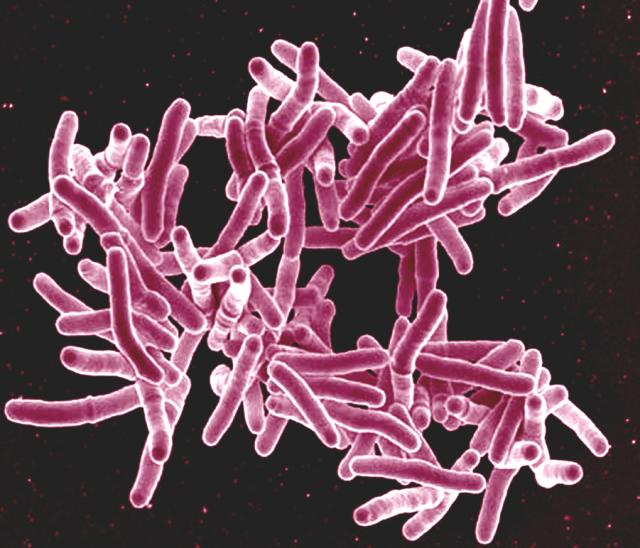
Much of Prof. Almqvist’s research is focused on tuberculosis, an infection that kills 1.6 million people every year. Some strains of the disease, such as Mycobacteria tuberculosis (Mtb), show formidable resistance to antibiotics, making them particularly difficult to treat.
However, Prof. Almqvist has joined forces with molecular biologist Christina Stallings at Washington University, US, to develop chemical compounds that break down the defences of these drug-resistant strains by inhibiting the formation of biofilm – a matrix of molecules that surrounds bacterial cells, providing protection against outside assaults. Enclosed in biofilm, bacteria can be up to 1,000-fold more resistant to antibiotics, but in the presence of Prof. Almqvist’s compounds, drug-resistant TB bacteria are rendered defenceless.
‘The tuberculosis bacteria are there and alive, but they don’t form a biofilm,’ said Prof. Almqvist. ‘With no biofilm, they get killed by the antibiotic.’
Prof. Almqvist says the compounds discovered in his lab – known as Mycobacterial Tolerance Inhibitors (MTIs) – show promise in two ways: they block Mtb’s defences against antibiotics, and they reduce treatment times for all TB patients from 6-9 months to just one month. His biochemical company, Quretech Bio, is now ready to start testing Mtb compounds on animal models.
Antibiotic-resistance breakers
Superbugs Methicillin-resistant Staphylococcus aureus (MRSA) and Vancomycin-resistant Enterococci (VRE) have also had their antibiotic resistance reversed in Prof Almqvist’s lab after exposure to antibiotic-resistance breakers (ARBs).
‘These are infections that can move to a very dramatic situation within hours if they are not treated effectively,’ said Prof. Almqvist.
He explains that ARBs could bring life-threatening infections under control by restoring the potency of antibiotics that have lost their edge. ‘We see this as a straightforward approach to a really big problem,’ he said.
As part of a project called QTB4AMR, he is developing a methodology for synthesising compounds that can then be tested and developed into ARBs. Compounds (some structurally similar to one another) are being evaluated for their ability to ‘strongly synergise’ with existing antibiotics at a rate of 100 to 500 new compounds per year.
‘We hope to motivate further development,’ said Prof. Almqvist, admitting that it will take ‘a lot of investment’ and ‘big engagement’ from both academic research bodies and pharmaceutical companies before his ARBs yield new treatments for patients.
To an extent, antibiotic resistance is a naturally occurring phenomenon. Bacteria mutate rapidly, and mutant strains that are able to survive a dose of antibiotics have an immediate evolutionary advantage. But our misuse and overuse of antibiotics is accelerating the process of resistance, putting us in a race to stay ahead of the superbugs. Resistance is a global catastrophe that kills 700,000 people every year. If solutions are not found fast, drug resistance is expected to cause 300 million premature deaths globally over the next 35 years.
Research aimed at finding new antibiotics has virtually stalled over the past 30 years. Of the products being tested today, only a handful are classed by the World Health Organisation (WHO) as innovative treatments with new modes of action that will add value to the arsenal of existing treatments (as opposed to reformulations of existing drugs). Most alarmingly, none of the new antibiotics in development are expected to be effective against the most dangerous forms of antibiotic-resistant bacteria.
Cost
So, why the lack of innovation? Engineering a new formulation is an expensive business, yet there’s pressure on doctors from health organisations around the world to prescribe antibiotics prudently. In other words, the world needs drugs that can sit on the shelf until they are absolutely essential – hardly an incentive for drug firms to invest billions in research (indeed, between 2003 and 2013, less than 5% of venture capital investment in pharmaceutical R&D was for antimicrobial development). What’s more, unlike many other life-saving medicines, antibiotics are generally used short-term and patients expect to pick them up cheaply.
‘We pay millions of euros for some therapies, but we expect antibiotics – which are life-saving drugs – to cost just a few euros,’ said Mariana Pinho, head of the bacterial cell biology laboratory at the Universidade Nova de Lisboa in Portugal.
She added: ‘This is why the idea of taking drugs that have already been approved for clinical use and repurposing them is very attractive. Re-sensitising bacteria is far more cost effective than developing new antibiotics.’
Like Prof. Almqvist, Prof. Pinho is on a quest to find ways to boost the performance of existing antibacterial medications. Though she stresses that the hunt for new antibiotics must never cease, she also believes in the potential of combination therapies, where washed-out antibiotics are restored to full working order after being blended with smart-acting new compounds.
Her ChronosAntibiotic project centres around Staphylococcus aureus, one of the most common multidrug-resistant bacterium in Europe. Staphylococcus aureus causes a raft of conditions, some of them serious, collectively known as staph infections.
Using super-resolution microscopy, she studies the mechanisms of the bacteria’s cell division, looking for moments in the cell cycle when the microbe is more susceptible to the actions of antibiotics. She believes that the addition of targeted resistance-busting compounds will allow antibiotics to use these windows of opportunity to best effect.
‘We want to find those weak points in the cell cycle,’ she said. ‘These are our long-term aims.’
Prof. Pinho is part of the team that has found a chink in the armour of MRSA drug resistance. A small molecule called DNAC-1, discovered by Prof. Pinho’s collaborators, has been seen to intensify the effect of the antibiotic oxacillin by damaging the microbe’s cell membrane. With the membrane disrupted, the antibiotic is free to stream in and kill the cell.
‘We see this as a straightforward approach to a really big problem.’
Prof. Fredrik Almqvist, Umeå University, Sweden
The cell membrane is just one of the potential targets for the action of new compounds. All cellular components and metabolic actions are coming under careful scrutiny for their susceptibility to damage and disruption.
To keep track of her lab’s findings, Prof. Pinho is building a library that pairs bacterial genes known as reporters with compounds that are found to inhibit the expression of these genes, thereby leaving the cell sensitive to antibiotics.
Her initial aim is to deepen the research community’s understanding of the cellular pathways that lead to drug resistance. ‘Just understanding the biology is an important start,’ she said.
By 2023, she hopes to have discovered reporters that are useful for antibiotics discovery to turn into a new drug or a smart compound to resurrect an existing one.
‘I would be very happy if, during the course of my life, I could contribute to the development of a new drug,’ she said.
Four bacterial defences against antibiotics
1. Invisibility cloak
The bacterial organism changes its physiology so that the antibiotic cannot find its target. Some bacteria will actively alter their cell wall structure to make it invisible.
2. Pump and flush
Bacteria regularly expel harmful compounds from their system using a protein to pump it out of the cell. These efflux pumps have evolved to now actively flush out antibiotics.
3. Protein shield
Proteins in the bacteria attach themselves to either antibiotics or the drug’s target within the bacterial cell. That protein then prevents the antibiotic from successfully binding to its target.
4. Counterattack
Some bacteria have evolved to produce defensive enzymes. They produce enzymes that either destroy or modify the antibiotic molecule and render them completely ineffective.
The research in this article was funded by the EU. If you liked this article, please consider sharing it on social media.
This post Can we reverse antibiotic resistance? was originally published on Horizon: the EU Research & Innovation magazine | European Commission.